Quantitative PCR
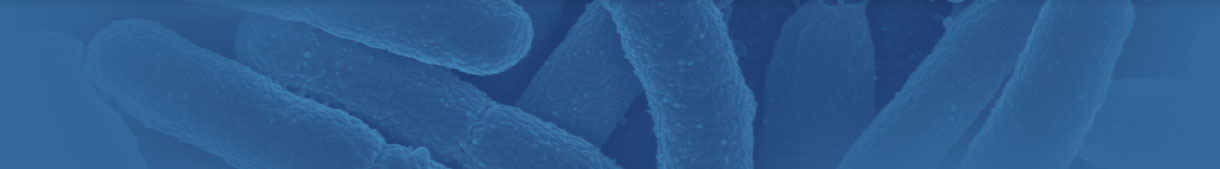
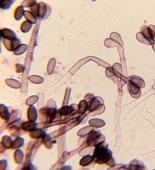 Mycologists traditionally rely on direct microscopic examination to arrive at the correct identifications of fungi. Since mycologists have identified over 100,000 different species of fungi, using traditional mycological methods can be a daunting task, requiring specialized training and years of experience. Culturing techniques require samples to be grown in a suitable medium, typically for one to two weeks for species identification. Speciation of molds is a skilled and labor-intensive task, even for the most experienced mycologists.
Mycologists traditionally rely on direct microscopic examination to arrive at the correct identifications of fungi. Since mycologists have identified over 100,000 different species of fungi, using traditional mycological methods can be a daunting task, requiring specialized training and years of experience. Culturing techniques require samples to be grown in a suitable medium, typically for one to two weeks for species identification. Speciation of molds is a skilled and labor-intensive task, even for the most experienced mycologists.
Research conducted at the US-EPA's National Exposure Research Laboratory has provided an alternative to traditional microscope-based mycology. Instead of samples being cultured for one or more weeks before results can be given, molds in both air and bulk samples can be speciated and quantified on a very rapid turnaround (one to two days) using quantitative real-time PCR (qPCR).
qPCR analysis has an instrument perform the identification and quantitation of the mold. Data is derived faster and can be more accurate with qPCR than with conventional culture-based methods. With qPCR, all the DNA present in a sample is extracted and is then identified and quantified using fluorescent-labeled probes that are specific to the mold species of interest.
Air samples are taken on a polycarbonate filter so overloading the sample is not an issue. Air volumes of 7000 L or more are common for qPCR samples, greatly increasing the sensitivity of air sampling when compared to Andersen and spore traps samples.
Bulk building material can also be submitted for analysis by qPCR when rapid speciation of observed mold growth is required. We can often provide results for a bulk sample on a same day turnaround.
qPCR has a limit of detection (LOD) of one spore per sample, but can still accurately quantify millions of spores per sample. In the instance of water intrusion molds such as Stachybotrys, Penicillium, and Aspergillus, accurate and rapid speciation and measurement are crucial to an effective risk assessment.
ERMI™
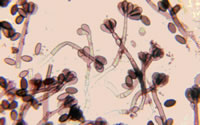 Fungal spores are a natural part of the outdoor environment. Spores are brought into buildings by air currents as well as being tracked in by people and animals. Once inside, spores can accumulate in dust, acting as a reservoir. Hidden mold problems due to water intrusion in a home can also lead to the accumulation of spores in household dust.
Fungal spores are a natural part of the outdoor environment. Spores are brought into buildings by air currents as well as being tracked in by people and animals. Once inside, spores can accumulate in dust, acting as a reservoir. Hidden mold problems due to water intrusion in a home can also lead to the accumulation of spores in household dust.
The Environmental Relative Moldiness Index (ERMI™) was developed by the US-EPA in 2002. They developed a nationwide database of molds present in both typical and mold-affected homes. Forensic Analytical Laboratories (FALI) was one of the initial laboratories to work with the US-EPA to analyze hundreds of dust extracts as part of the Department of Housing and Urban Development's (HUD) American Healthy Homes Survey in order to validate this procedure. The ERMI™ aims to reduce the effect of variables such as geography, construction, and housekeeping when comparing the homes with molds associated with water intrusion to homes with molds that naturally occur in "healthy" homes.
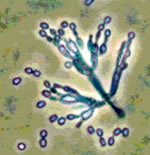 The ERMI™ is designed to objectively rank the 'relative moldiness' of a given home. Using a panel of 36 primer-probe sets, the concentrations of molds naturally present in healthy homes are statistically compared in the concentrations of molds in typical mold-affected homes. Based on this comparison, an ERMI™ Score is calculated. This score shows your home's position on the US-EPA's Relative Moldiness Index – the probability that your home has been affected in the recent past or is presently affected by a mold growth event. It can help make the decision on how to proceed to decrease potential health risks associated with mold.
The ERMI™ is designed to objectively rank the 'relative moldiness' of a given home. Using a panel of 36 primer-probe sets, the concentrations of molds naturally present in healthy homes are statistically compared in the concentrations of molds in typical mold-affected homes. Based on this comparison, an ERMI™ Score is calculated. This score shows your home's position on the US-EPA's Relative Moldiness Index – the probability that your home has been affected in the recent past or is presently affected by a mold growth event. It can help make the decision on how to proceed to decrease potential health risks associated with mold.
FALI is happy to offer ERMI™ testing as one of our ten customized PCR panels. Since ERMI™ results quantify the 36 species of molds most commonly found indoors by qPCR, you get the best of both worlds: an ERMI™ score that quantifies your home's position on the Relative Moldiness Index, plus quantification of all 36 species that may be present.